Jigsaw Algorithm Library is Publicly Available
We are pleased to announce that we have released our algorithm library to the public at https://public.jigsaw.io. This is something we have wanted to do for years, but there was always something else that we wanted to include or change. In the spirit of “perfect is the enemy of good”, we decided that the algorithm library was finally “good enough” to make it available to others. It is still a work in progress. We hope researchers who conduct studies using healthcare data find it useful.
Why put this out there?
We have spent many years struggling with the same issues that all researchers face when trying to extract information from healthcare data. Algorithms are hard to find, poorly documented and difficult to implement consistently. It is time to solve the problem.
Who is funding this?
This project is completely self-funded. Please keep in mind that we have limited resources, and we may not be able to accommodate requests. It turns out that it is surprisingly challenging to make something publicly and freely available, so we appreciate your understanding.
Collaboration
If any organization is interested in collaborating with us on algorithms, we welcome it – even from organizations that might be considered “competitors”. Algorithms should be freely available and compatible with other systems than our own. We are willing to see if we can make them work with other systems, even proprietary ones.
Feedback
We welcome suggestions. We are happy to consider algorithm modifications if an algorithm can be improved in any way. We would like our algorithms to be usable by anyone. Along these lines, we are willing to consider adding other algorithms to our library, as long they can be made public.
Browser requirements
The algorithm library is designed and tested against Google Chrome because of its popularity. Other browsers may work, but we don’t provide cross-browser support at this time.
But wait, there’s more!
In addition to the high-level details above, we also provide more detailed technical information and limitations below, as well as an example algorithm.
Technical Details
What is an algorithm library?
Like most libraries, the Jigsaw algorithm library stores algorithms, facilitates searching for algorithms, and contains tools for creating, editing, and versioning algorithms. The library enables algorithm sharing in two ways: as a stand-alone HTML file downloaded from the website, or via the algorithm’s unique URL.
How to use the Jigsaw algorithm library
Anyone interested can simply navigate to the algorithm section of the Jigsaw application to find algorithms. Users can then search for algorithms by different characteristics. Clicking on any algorithm’s name opens it, and the user is presented with algorithm attributes as well as a diagram of the algorithm specification. Users can click on the diagram to get more detailed information about the specifications.
We cover some of the basics below, and we will provide more granular instructions in the future.
What is an algorithm?
An algorithm has two parts – attributes and specifications.
Algorithm attributes include all of the information that makes algorithms friendly to researchers. This includes titles, tags, documentation, variable names, and even evidence of validity (e.g., sensitivity and specificity).
Algorithm specifications are comprised of the code(s) and operator(s) that are needed to identify information for an analysis dataset. The most basic operation of an algorithm is to “select” records with the code(s) of interest. This means that the simplest possible algorithm specification is simply to select all records using a single code (see Standardization for more on codes). However, some algorithms can have thousands of codes. Algorithms can also require operations related to the context in which the code was used (provenance), have temporal specifications (before, after, within, etc.), require relationships to other codes (co-reported), use filters (first, last, etc.), or specify other operations.
Sometimes algorithm specifications require many operations in sequence. For these use cases, we create special operators to make them easier to implement. The best example is an operator that looks for a code that occurs at least 1 time in the inpatient setting or at least 2 times in the outpatient setting within specified time intervals (sometimes referred to as a “one inpatient or two outpatient” operation). These operators make creating algorithm specifications much simpler.
What does an algorithm specification produce?
An algorithm specification is a representation of a table that is created and, potentially, modified. When implemented against data, every algorithm starts with a selection operator to create the first table based on the codes in the specification. This table can then be passed to subsequent operators for additional modification. The output of an algorithm is a table that includes all of the relevant records for each person as modified by the specified operations. As an example, a specification could first select all records with at least one diabetes code based on a list of diabetes codes, and then filter these records to retain only the first code for each person.
The core of each table includes a person id, details about the exact table and record that was returned, the start and end dates, the vocabulary (e.g., ICD-10-CM), and the specific code. Additional columns are dynamically added for selected types of records (e.g., prescription records will add additional columns specific to prescriptions, like dosing information).
How algorithms operate
We have defined all algorithm operations that are commonly used for research using ConceptQL (see ConceptQL blog post). Within the algorithm library, there is a hyperlink from each operator to our ConceptQL specification in Github that provides a more detailed description of how it functions.
Implicitly, all algorithm operations are conducted by person since almost all research is conducted on cohorts of people. All algorithms are read left to right, and only one table is ever produced at the end of any operation. This is true even if multiple tables are used as inputs to an operation. For algorithms with two sides to them, the left side takes the input of the previous operation and modifies it using information from the right side. Tables on the right side are never passed through and included in the output.
If the above doesn’t make any sense, don’t worry. We will provide more details about how algorithms operate in a future post.
Visual representation
We have found that a visual representation of an algorithm specification is extremely helpful for communicating it to others. As a result, an algorithm is specified using a visual algorithm builder that generates ConceptQL statements and encapsulates the algorithm specification. See below for an example.
Algorithm scope
Do we have every possible operation that might ever be needed? Probably not. Hence, it is possible to extend Jigsaw with new operators as the need arises. However, we expect the need for new operators to be relatively rare. Many times, the desired output requires some creativity using existing operations.
Alternatively, it may be that the desired operation is really a protocol specification, and not an algorithm specification. It is beyond this blog post to define the difference between an algorithm specification and a protocol specification precisely. However, in general, an algorithm specifies the variable of interest, and the protocol defines the context in which the algorithm operates. The separation between an algorithm and a protocol ensures that an algorithm is as reusable as possible.
To make this a bit more concrete, an algorithm might define a myocardial infarction. Or it might define diabetes diagnosed at least 30 days before a myocardial infarction. However, a myocardial infarction within 30 days of the index date, or as an inclusion or exclusion criterion, or as a censoring variable – those are ways of using a myocardial infarction algorithm in the context of a protocol. The same idea applies with clean periods or lookback periods – those are protocol-specific ideas that are not part of an algorithm’s scope.
As a side note, users may see some algorithms in our library that violate this principle. This is because our thinking about the line between algorithms and protocols has evolved over the years. As a result, we have added features to our protocol builder to help keep the distinction as clear as possible. But since we don’t readily remove algorithms we have used, these algorithms remain in the algorithm library.
It isn’t clear how algorithms are used to create analysis data sets
Protocols, not algorithms, specify the analysis datasets that need to be created. Protocols specify the index date, inclusion and exclusion criteria, baseline variables, lookback periods, clean periods, date restrictions, and outcome variables. Protocols use algorithms to specify each variable within the context of a protocol. It is much like assembling a jigsaw puzzle (protocol) using pieces (algorithms). Hence, our software is named “Jigsaw”.
As a simple example, the specification for myocardial infarction can be identical regardless of whether it is needed as a baseline variable, index event, or outcome. However, different records will be selected depending on which context required.
To illustrate how algorithms fit into protocols, we have made our entire Jigsaw application available, including our protocol builder, albeit in read-only form. There is a demonstration protocol available within the application.
Limitations
Disclaimer
While we built Jigsaw and the algorithms carefully, we make no guarantees of correctness or suitability for a specific study. Jigsaw is a tool to make research easier and more transparent; however, it still requires that users fully understand all aspects of the research they are conducting, their data source(s), and the limitations of real-world data.
Is every algorithm ready for research?
The algorithm library contains over 1,000 algorithms and spans many years of development and growth. Therefore, not every algorithm is ready to be used in its current state. Some were created as examples, some were created prior to ICD-10-CM data becoming available, and some were created or updated very recently. Most algorithms are classified as “Draft” even though they have been extensively used. Some “Finalized” algorithms should probably be updated. As mentioned above, this is a work in progress and we only update algorithms as we reuse them. We suggest looking at the date of the most recent update to get a sense of how current an algorithm is. Users should do their own due-diligence before using any algorithm.
Is an algorithm specific to a data model or software system?
The algorithm library is agnostic to both the data structure and the software used for implementation. The algorithm simply captures the specifications for an algorithm. For example, an algorithm can encapsulate the idea that we are looking for a “diabetes diagnosis at least 30 days before a myocardial infarction”.
As such, it is not written in SQL, SAS, R, Python, or any language that can be used directly on data. Instead, it is specified in JSON using the ConceptQL language. Jigsaw uses the specification to dynamically generate SQL and implement the algorithm against data. In theory, ConceptQL could generate something other than SQL, but practically, SQL is so widely available there has been no reason to consider other options. This approach allows us to generate SQL that is specific to the idiosyncrasies of different database implementations of SQL. For example, we can generate SQL specific to PostgreSQL, Impala, SQLite or most other database systems that follow SQL standards.
Furthermore, we can generate SQL that works directly against different data models, like the Generalized Data Model (GDM), OMOP, PCORnet, and Sentinel. (Note that support for OMOP, PCORnet, and Sentinel is not currently implemented because we have not had a use case for them.)
To see what the SQL code looks like for an algorithm, users can dynamically generate the necessary SQL from inside the algorithm diagram. The SQL is specific to the Generalized Data Model, and it produces PostgreSQL queries. See below for a complete example of an algorithm that includes the diagram, attributes, ConceptQL JSON statement, and the resulting SQL.
Why not make the entire application available?
While Jigsaw is copyrighted and remains our intellectual property, our hope is that we can make its full functionality available to other researchers. To do that, we must make ETL easier first, which is not a trivial issue. We have made progress on that front as well, and are willing to collaborate with organizations interested in adopting the Generalized Data Model and/or Jigsaw.
Example Algorithm
Below is an example algorithm that identifies diabetes diagnoses coming at least 30 days prior to a myocardial infarction. Both ICD-9-CM and ICD-10-CM codes are used to identify diabetes and myocardial infarction. Note that this is an example and may not represent the correct codes or a clinically meaningful event.
Algorithm diagram
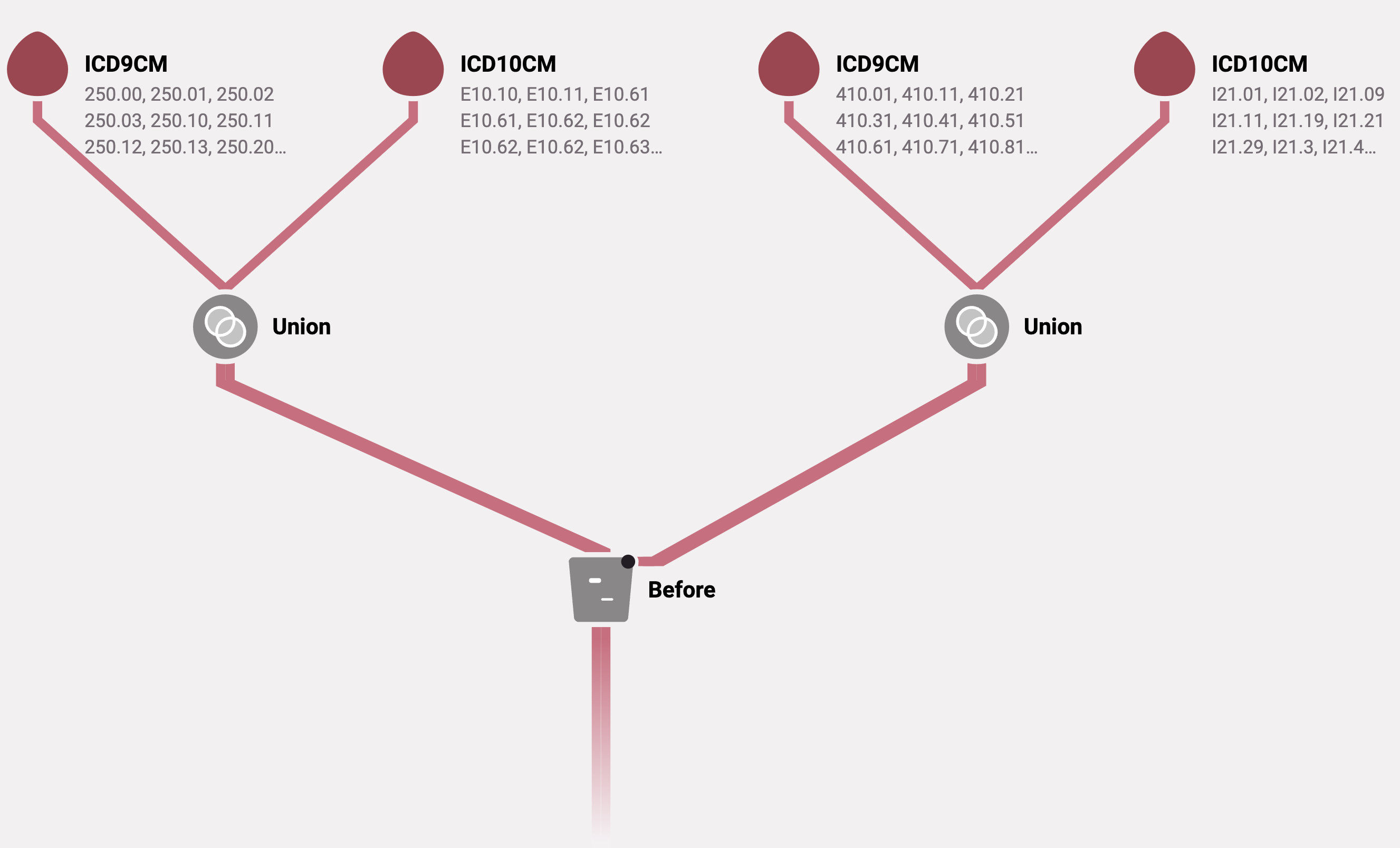
Algorithm attributes
ConceptQL
Below is the ConceptQL statement that corresponds to the diagram above.
[
"before",
{
"left": [
"union",
[
"icd9cm",
"250.00",
"250.01",
"250.02",
"250.03",
"250.10",
"250.11",
"250.12",
"250.13",
"250.20",
"250.21",
"250.22",
"250.23",
"250.30",
"250.31",
"250.32",
"250.33",
"250.80",
"250.81",
"250.82",
"250.83",
"250.90",
"250.91",
"250.92",
"250.93"
],
[
"icd10cm",
"E10.10",
"E10.11",
"E10.61",
"E10.61",
"E10.62",
"E10.62",
"E10.62",
"E10.62",
"E10.63",
"E10.63",
"E10.64",
"E10.64",
"E10.65",
"E10.69",
"E10.8",
"E10.9",
"E11.00",
"E11.01",
"E11.10",
"E11.11",
"E11.61",
"E11.61",
"E11.62",
"E11.62",
"E11.62",
"E11.62",
"E11.63",
"E11.63",
"E11.64",
"E11.64",
"E11.65",
"E11.69",
"E11.8",
"E11.9",
"E13.00",
"E13.01",
"E13.10",
"E13.11",
"E13.61",
"E13.61",
"E13.62",
"E13.62",
"E13.62",
"E13.62",
"E13.63",
"E13.63",
"E13.64",
"E13.64",
"E13.65",
"E13.69",
"E13.8",
"E13.9"
]
],
"right": [
"union",
[
"icd9cm",
"410.01",
"410.11",
"410.21",
"410.31",
"410.41",
"410.51",
"410.61",
"410.71",
"410.81",
"410.91"
],
[
"icd10cm",
"I21.01",
"I21.02",
"I21.09",
"I21.11",
"I21.19",
"I21.21",
"I21.29",
"I21.3",
"I21.4",
"I21.9",
"I21.A1",
"I21.A9",
"I22.0",
"I22.1",
"I22.2",
"I22.8",
"I22.9"
]
],
"at_least": "30d"
}
]
SQL
The code below is generated from the ConceptQL specification. This particular SQL code will create an output table from data stored in a PostgreSQL relational database based on the Generalized Data Model schema.
SELECT
*
FROM ( WITH "before_22_3_24a2b6fb9b0f37c3735c654d1ffd3cab" AS MATERIALIZED (
SELECT
*
FROM (
SELECT
"person_id",
"criterion_id",
"criterion_table",
"criterion_domain",
"start_date",
"end_date",
"source_value",
"source_vocabulary_id",
"label"
FROM (
SELECT
*
FROM (
SELECT
*
FROM (
SELECT
*
FROM (
WITH "union_22_2_64ac7676c653ae02d50931398ee95903" AS MATERIALIZED (
SELECT
*
FROM (
SELECT
"person_id",
"criterion_id",
"criterion_table",
"criterion_domain",
"start_date",
"end_date",
"source_value",
"source_vocabulary_id",
"label"
FROM (
SELECT
"person_id",
"criterion_id",
"criterion_table",
"criterion_domain",
"start_date",
"end_date",
"source_value",
"source_vocabulary_id",
"label"
FROM (
SELECT
*
FROM (
SELECT
"patient_id" AS "person_id",
"id" AS "criterion_id",
CAST(
'clinical_codes' AS text
) AS "criterion_table",
CAST(
'condition_occurrence' AS text
) AS "criterion_domain",
"start_date",
"end_date",
CAST(
"clinical_code_source_value" AS text
) AS "source_value",
CAST(
"clinical_code_vocabulary_id" AS text
) AS "source_vocabulary_id",
CAST(
NULL AS text
) AS "label"
FROM
"clinical_codes"
WHERE ((
"clinical_code_concept_id" IN (
SELECT
"id"
FROM
"concepts"
WHERE ((
"vocabulary_id" = 'ICD9CM'
)
AND (
"concept_code" IN (
'250.00', '250.01', '250.02', '250.03', '250.10', '250.11', '250.12', '250.13', '250.20', '250.21', '250.22', '250.23', '250.30', '250.31', '250.32', '250.33', '250.80', '250.81', '250.82', '250.83', '250.90', '250.91', '250.92', '250.93'
)
)
)
)
)
OR (
"clinical_code_concept_id" IN (
SELECT
"id"
FROM
"concepts"
WHERE ((
"vocabulary_id" = 'ICD10CM'
)
AND (
"concept_code" IN (
'E10.10', 'E10.11', 'E10.61', 'E10.61', 'E10.62', 'E10.62', 'E10.62', 'E10.62', 'E10.63', 'E10.63', 'E10.64', 'E10.64', 'E10.65', 'E10.69', 'E10.8', 'E10.9', 'E11.00', 'E11.01', 'E11.10', 'E11.11', 'E11.61', 'E11.61', 'E11.62', 'E11.62', 'E11.62', 'E11.62', 'E11.63', 'E11.63', 'E11.64', 'E11.64', 'E11.65', 'E11.69', 'E11.8', 'E11.9', 'E13.00', 'E13.01', 'E13.10', 'E13.11', 'E13.61', 'E13.61', 'E13.62', 'E13.62', 'E13.62', 'E13.62', 'E13.63', 'E13.63', 'E13.64', 'E13.64', 'E13.65', 'E13.69', 'E13.8', 'E13.9'
)
)
)
)
)
) -- #<ConceptQL::Operator::icd9cm @arguments=["250.00", "250.01", "250.02", "250.03", "250.10", "250.11", "250.12", "250.13", "250.20", "250.21", "250.22", "250.23", "250.30", "250.31", "250.32", "250.33", "250.80", "250.81", "250.82", "250.83", "250.90", "250.91", "250.92", "250.93"]>
) AS "t1"
) AS "t1"
) AS "t1" -- #<ConceptQL::Operators::Union @upstreams=[#<ConceptQL::Operator::icd9cm @arguments=["250.00", "250.01", "250.02", "250.03", "250.10", "250.11", "250.12", "250.13", "250.20", "250.21", "250.22", "250.23", "250.30", "250.31", "250.32", "250.33", "250.80", "250.81", "250.82", "250.83", "250.90", "250.91", "250.92", "250.93"]>, #<ConceptQL::Operator::icd10cm @arguments=["E10.10", "E10.11", "E10.61", "E10.61", "E10.62", "E10.62", "E10.62", "E10.62", "E10.63", "E10.63", "E10.64", "E10.64", "E10.65", "E10.69", "E10.8", "E10.9", "E11.00", "E11.01", "E11.10", "E11.11", "E11.61", "E11.61", "E11.62", "E11.62", "E11.62", "E11.62", "E11.63", "E11.63", "E11.64", "E11.64", "E11.65", "E11.69", "E11.8", "E11.9", "E13.00", "E13.01", "E13.10", "E13.11", "E13.61", "E13.61", "E13.62", "E13.62", "E13.62", "E13.62", "E13.63", "E13.63", "E13.64", "E13.64", "E13.65", "E13.69", "E13.8", "E13.9"]>]>
) AS "t1"
)
SELECT
*
FROM
"union_22_2_64ac7676c653ae02d50931398ee95903"
) AS "l"
) AS "l"
WHERE (
EXISTS (
SELECT
1
FROM (
SELECT
*
FROM (
SELECT
"person_id",
max(
"start_date"
) AS "start_date"
FROM (
WITH "union_22_1_d3d7bbd424bca0fe3b3f966d4ee80692" AS MATERIALIZED (
SELECT
*
FROM (
SELECT
"person_id",
"criterion_id",
"criterion_table",
"criterion_domain",
"start_date",
"end_date",
"source_value",
"source_vocabulary_id",
"label"
FROM (
SELECT
"person_id",
"criterion_id",
"criterion_table",
"criterion_domain",
"start_date",
"end_date",
"source_value",
"source_vocabulary_id",
"label"
FROM (
SELECT
*
FROM (
SELECT
"patient_id" AS "person_id",
"id" AS "criterion_id",
CAST(
'clinical_codes' AS text
) AS "criterion_table",
CAST(
'condition_occurrence' AS text
) AS "criterion_domain",
"start_date",
"end_date",
CAST(
"clinical_code_source_value" AS text
) AS "source_value",
CAST(
"clinical_code_vocabulary_id" AS text
) AS "source_vocabulary_id",
CAST(
NULL AS text
) AS "label"
FROM
"clinical_codes"
WHERE ((
"clinical_code_concept_id" IN (
SELECT
"id"
FROM
"concepts"
WHERE ((
"vocabulary_id" = 'ICD9CM'
)
AND (
"concept_code" IN (
'410.01', '410.11', '410.21', '410.31', '410.41', '410.51', '410.61', '410.71', '410.81', '410.91'
)
)
)
)
)
OR (
"clinical_code_concept_id" IN (
SELECT
"id"
FROM
"concepts"
WHERE ((
"vocabulary_id" = 'ICD10CM'
)
AND (
"concept_code" IN (
'I21.01', 'I21.02', 'I21.09', 'I21.11', 'I21.19', 'I21.21', 'I21.29', 'I21.3', 'I21.4', 'I21.9', 'I21.A1', 'I21.A9', 'I22.0', 'I22.1', 'I22.2', 'I22.8', 'I22.9'
)
)
)
)
)
) -- #<ConceptQL::Operator::icd9cm @arguments=["410.01", "410.11", "410.21", "410.31", "410.41", "410.51", "410.61", "410.71", "410.81", "410.91"]>
) AS "t1"
) AS "t1"
) AS "t1" -- #<ConceptQL::Operators::Union @upstreams=[#<ConceptQL::Operator::icd9cm @arguments=["410.01", "410.11", "410.21", "410.31", "410.41", "410.51", "410.61", "410.71", "410.81", "410.91"]>, #<ConceptQL::Operator::icd10cm @arguments=["I21.01", "I21.02", "I21.09", "I21.11", "I21.19", "I21.21", "I21.29", "I21.3", "I21.4", "I21.9", "I21.A1", "I21.A9", "I22.0", "I22.1", "I22.2", "I22.8", "I22.9"]>]>
) AS "t1"
)
SELECT
*
FROM
"union_22_1_d3d7bbd424bca0fe3b3f966d4ee80692"
) AS "t1"
GROUP BY
"person_id"
) AS "r"
) AS "r"
WHERE ((
"l"."person_id" = "r"."person_id"
)
AND (
"l"."end_date" < CAST((
CAST(
"r"."start_date" AS timestamp
) + make_interval(
days := - 30
)
) AS date
)
)
)
)
)
) AS "t1"
) AS "t1" -- #<ConceptQL::Operators::Before @upstreams={:left=> #<ConceptQL::Operators::Union @upstreams=[#<ConceptQL::Operator::icd9cm @arguments=["250.00", "250.01", "250.02", "250.03", "250.10", "250.11", "250.12", "250.13", "250.20", "250.21", "250.22", "250.23", "250.30", "250.31", "250.32", "250.33", "250.80", "250.81", "250.82", "250.83", "250.90", "250.91", "250.92", "250.93"]>, #<ConceptQL::Operator::icd10cm @arguments=["E10.10", "E10.11", "E10.61", "E10.61", "E10.62", "E10.62", "E10.62", "E10.62", "E10.63", "E10.63", "E10.64", "E10.64", "E10.65", "E10.69", "E10.8", "E10.9", "E11.00", "E11.01", "E11.10", "E11.11", "E11.61", "E11.61", "E11.62", "E11.62", "E11.62", "E11.62", "E11.63", "E11.63", "E11.64", "E11.64", "E11.65", "E11.69", "E11.8", "E11.9", "E13.00", "E13.01", "E13.10", "E13.11", "E13.61", "E13.61", "E13.62", "E13.62", "E13.62", "E13.62", "E13.63", "E13.63", "E13.64", "E13.64", "E13.65", "E13.69", "E13.8", "E13.9"]>]>, :right=> #<ConceptQL::Operators::Union @upstreams=[#<ConceptQL::Operator::icd9cm @arguments=["410.01", "410.11", "410.21", "410.31", "410.41", "410.51", "410.61", "410.71", "410.81", "410.91"]>, #<ConceptQL::Operator::icd10cm @arguments=["I21.01", "I21.02", "I21.09", "I21.11", "I21.19", "I21.21", "I21.29", "I21.3", "I21.4", "I21.9", "I21.A1", "I21.A9", "I22.0", "I22.1", "I22.2", "I22.8", "I22.9"]>]>}>
) AS "t1"
)
SELECT
*
FROM
"before_22_3_24a2b6fb9b0f37c3735c654d1ffd3cab") AS "t1"
 by Outcomes Insights, Inc.
by Outcomes Insights, Inc.